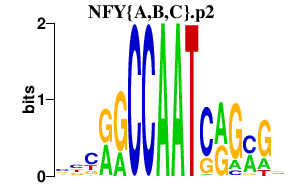
|
chr6_+_26045611
|
4.537
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr6_+_26199712
|
2.848
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr19_+_38893766
|
2.688
|
NM_174905
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr1_+_151763174
|
2.246
|
|
|
|
|
chr17_-_80656461
|
1.827
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr6_-_26043884
|
1.644
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr17_+_46019022
|
1.530
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr19_-_49140569
|
1.402
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr1_+_227127937
|
1.332
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_227127999
|
1.315
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chrX_+_49047887
|
1.296
|
|
|
|
|
chr4_+_88720701
|
1.291
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr11_-_67275549
|
1.278
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr3_-_27410852
|
1.256
|
NM_199347
|
NEK10
|
NIMA (never in mitosis gene a)- related kinase 10
|
|
chr1_+_227127959
|
1.248
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr11_-_45939361
|
1.240
|
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr17_+_46018888
|
1.226
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr17_+_48172508
|
1.216
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr19_-_2944894
|
1.208
|
NM_021217
|
ZNF77
|
zinc finger protein 77
|
|
chr16_+_72698996
|
1.190
|
|
|
|
|
chr1_+_227127993
|
1.186
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr4_-_120375770
|
1.177
|
|
|
|
|
chr10_-_15130773
|
1.171
|
NM_001039844
|
ACBD7
|
acyl-CoA binding domain containing 7
|
|
chr9_-_34329151
|
1.161
|
NM_194313
|
KIF24
|
kinesin family member 24
|
|
chr17_-_8059637
|
1.104
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr2_-_240322692
|
1.101
|
|
HDAC4
|
histone deacetylase 4
|
|
chr7_+_74988447
|
1.096
|
|
STAG3L3
STAG3L2
STAG3L1
|
stromal antigen 3-like 3
stromal antigen 3-like 2
stromal antigen 3-like 1
|
|
chr22_+_42486904
|
1.092
|
|
LOC100132273
|
uncharacterized LOC100132273
|
|
chr15_-_23378167
|
1.090
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr17_-_7197896
|
1.082
|
|
YBX2
|
Y box binding protein 2
|
|
chr19_+_14640413
|
1.069
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr19_-_55677919
|
1.066
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr4_+_156680125
|
1.044
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr2_-_240322620
|
1.043
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr19_+_38893804
|
1.042
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chrX_+_52926334
|
1.025
|
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr17_-_7197811
|
1.014
|
|
YBX2
|
Y box binding protein 2
|
|
chr16_-_4852571
|
1.007
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr19_+_19174792
|
0.982
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr21_-_47743753
|
0.968
|
NM_058180
|
C21orf58
|
chromosome 21 open reading frame 58
|
|
chr1_-_150602020
|
0.953
|
|
ENSA
|
endosulfine alpha
|
|
chr21_-_34852282
|
0.948
|
NM_006134
|
TMEM50B
|
transmembrane protein 50B
|
|
chr6_-_26197477
|
0.945
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr17_+_77071002
|
0.940
|
NM_001042573
|
ENGASE
|
endo-beta-N-acetylglucosaminidase
|
|
chr1_-_16161279
|
0.934
|
|
|
|
|
chr12_-_6665220
|
0.926
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr1_-_6052481
|
0.912
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr1_-_28241233
|
0.899
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr19_+_14640363
|
0.884
|
NM_138501
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chrX_+_52926358
|
0.883
|
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr3_-_59035700
|
0.873
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr19_+_14640406
|
0.870
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr16_+_103892
|
0.867
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr7_+_148982371
|
0.863
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr16_-_1661969
|
0.860
|
NM_014714
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas)
|
|
chr6_-_26033795
|
0.858
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr17_-_8198605
|
0.854
|
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr6_-_27835308
|
0.849
|
NM_005322
|
HIST1H1B
|
histone cluster 1, H1b
|
|
chr21_-_34852226
|
0.849
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr2_-_239148626
|
0.847
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr8_+_118532953
|
0.842
|
NM_080651
|
MED30
|
mediator complex subunit 30
|
|
chr11_+_64879294
|
0.841
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr6_-_32095974
|
0.837
|
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr19_+_36266366
|
0.834
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr19_+_13001966
|
0.827
|
NM_000159
NM_013976
|
GCDH
|
glutaryl-CoA dehydrogenase
|
|
chr1_+_64239566
|
0.822
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr1_+_145438437
|
0.795
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr1_+_43312288
|
0.794
|
|
|
|
|
chr4_-_1722776
|
0.793
|
|
TMEM129
|
transmembrane protein 129
|
|
chr2_-_73460326
|
0.790
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chr1_+_192778168
|
0.773
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr10_+_96522457
|
0.772
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr6_-_32095993
|
0.771
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr6_-_27858569
|
0.769
|
NM_003535
|
HIST1H3J
|
histone cluster 1, H3j
|
|
chr14_+_29236171
|
0.768
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr17_-_7197866
|
0.760
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr17_+_46048406
|
0.758
|
NM_176096
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr4_-_7044664
|
0.756
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr16_-_74455239
|
0.755
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr12_-_56727720
|
0.746
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr17_-_35293920
|
0.745
|
|
|
|
|
chr14_+_53019865
|
0.744
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr14_+_105941061
|
0.741
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chrX_+_49126326
|
0.740
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr20_-_50418946
|
0.736
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr14_+_70346113
|
0.736
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr14_-_75389901
|
0.736
|
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr2_+_150187028
|
0.730
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr4_-_775586
|
0.724
|
|
LOC100129917
|
uncharacterized LOC100129917
|
|
chr17_-_7307413
|
0.722
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 readthrough
chromosome 17 open reading frame 61
|
|
chr17_+_1619816
|
0.718
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr2_+_73144603
|
0.716
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chr19_-_14228560
|
0.713
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr17_-_56595192
|
0.710
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr11_+_34460421
|
0.706
|
NM_001752
|
CAT
|
catalase
|
|
chr6_+_31783314
|
0.703
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr14_-_90421032
|
0.693
|
NM_145231
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chr16_-_1876528
|
0.690
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr20_+_13976263
|
0.688
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr19_-_14606939
|
0.685
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr7_+_100209724
|
0.685
|
NM_001040097
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr16_+_8768403
|
0.679
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr9_-_128003615
|
0.678
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr11_-_45939568
|
0.677
|
NM_004813
NM_057174
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr1_-_23857284
|
0.664
|
|
E2F2
|
E2F transcription factor 2
|
|
chr16_+_1833003
|
0.660
|
|
|
|
|
chr1_-_149783910
|
0.660
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr16_+_103859
|
0.656
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr7_-_65215903
|
0.655
|
|
|
|
|
chr16_+_4743693
|
0.653
|
NM_001193452
NM_032349
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr7_+_99070428
|
0.653
|
|
ZNF789
|
zinc finger protein 789
|
|
chr14_-_24584125
|
0.651
|
|
NRL
|
neural retina leucine zipper
|
|
chr13_-_67804432
|
0.648
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr6_-_26271588
|
0.648
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr6_+_33172418
|
0.646
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr1_-_117664349
|
0.645
|
NM_001145635
NM_025188
|
TRIM45
|
tripartite motif containing 45
|
|
chr6_+_27114860
|
0.644
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr12_+_117176085
|
0.643
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr5_-_107717798
|
0.642
|
NM_001163315
|
FBXL17
|
F-box and leucine-rich repeat protein 17
|
|
chr3_-_169482847
|
0.641
|
|
TERC
|
telomerase RNA component
|
|
chr4_-_1685717
|
0.640
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr1_+_43148096
|
0.639
|
|
YBX1
|
Y box binding protein 1
|
|
chr19_-_14247400
|
0.638
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr19_+_50031232
|
0.638
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr15_-_64673569
|
0.637
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr17_-_58212896
|
0.636
|
|
|
|
|
chr17_-_43025027
|
0.630
|
NM_001080443
|
KIF18B
|
kinesin family member 18B
|
|
chr1_-_44173010
|
0.630
|
|
LOC100132774
|
uncharacterized LOC100132774
|
|
chr3_-_120461350
|
0.629
|
|
|
|
|
chr12_-_54982271
|
0.624
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr1_-_6052410
|
0.622
|
|
NPHP4
|
nephronophthisis 4
|
|
chr11_+_66406189
|
0.620
|
|
RBM4
|
RNA binding motif protein 4
|
|
chr1_-_36915969
|
0.618
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr3_+_50229022
|
0.618
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr22_+_21986968
|
0.617
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr5_+_108084638
|
0.616
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr5_-_149792276
|
0.614
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr3_-_186288331
|
0.612
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr9_+_132083294
|
0.611
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr6_+_99282597
|
0.608
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr10_+_99344421
|
0.607
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr19_+_8455265
|
0.607
|
|
RAB11B
|
RAB11B, member RAS oncogene family
|
|
chr10_-_71906452
|
0.603
|
NM_001040273
NM_173555
|
TYSND1
|
trypsin domain containing 1
|
|
chr21_-_47878592
|
0.601
|
|
|
|
|
chr7_+_98246595
|
0.600
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr1_-_21671905
|
0.599
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_-_71454200
|
0.597
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr12_-_56727446
|
0.594
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr17_+_61627841
|
0.593
|
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr4_-_84035908
|
0.591
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr1_-_149858114
|
0.590
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr16_-_2723374
|
0.587
|
|
ERVK13-1
|
endogenous retrovirus group K13, member 1
|
|
chr15_-_20711386
|
0.583
|
|
HERC2P3
|
hect domain and RLD 2 pseudogene 3
|
|
chr6_+_31783280
|
0.581
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr2_-_128784736
|
0.581
|
|
SAP130
|
Sin3A-associated protein, 130kDa
|
|
chr3_-_37034567
|
0.579
|
|
|
|
|
chr12_+_49716970
|
0.579
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr6_+_31783530
|
0.579
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr16_-_68482373
|
0.577
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr2_+_177053306
|
0.576
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr17_-_3539384
|
0.575
|
NM_013276
|
SHPK
|
sedoheptulokinase
|
|
chr19_-_14606893
|
0.574
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr7_-_65235688
|
0.573
|
|
LOC441242
|
uncharacterized LOC441242
|
|
chr20_-_32274154
|
0.571
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr12_+_7013896
|
0.569
|
NM_001135217
NM_006992
NM_201650
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr19_+_7694658
|
0.568
|
NM_001171155
|
C19orf79
|
chromosome 19 open reading frame 79
|
|
chr17_+_42634598
|
0.566
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr17_-_76183074
|
0.565
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr17_-_8113862
|
0.561
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr1_+_16062808
|
0.560
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr1_-_149859465
|
0.560
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr16_-_30773303
|
0.558
|
NM_001014979
NM_001195620
|
C16orf93
|
chromosome 16 open reading frame 93
|
|
chr10_-_131909061
|
0.556
|
|
LOC387723
|
uncharacterized LOC387723
|
|
chr14_-_74551073
|
0.555
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr21_-_34852163
|
0.555
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr20_-_60982334
|
0.552
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr11_-_47198379
|
0.551
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr11_+_61891414
|
0.550
|
NM_001040694
NM_020238
|
INCENP
|
inner centromere protein antigens 135/155kDa
|
|
chr3_+_111393522
|
0.549
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr4_-_174255454
|
0.549
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chrX_+_49126299
|
0.548
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr1_-_28240953
|
0.545
|
|
RPA2
|
replication protein A2, 32kDa
|
|
chr7_+_74306895
|
0.544
|
|
PMS2P5
|
postmeiotic segregation increased 2 pseudogene 5
|
|
chr10_-_71906409
|
0.543
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr17_+_61627976
|
0.541
|
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr19_+_14640403
|
0.538
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr12_-_53448167
|
0.538
|
|
LOC283335
|
uncharacterized LOC283335
|
|
chr17_-_8055684
|
0.533
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr22_+_38054701
|
0.532
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr7_+_99070513
|
0.526
|
NM_001013258
NM_213603
|
ZNF789
|
zinc finger protein 789
|
|
chr5_-_149535420
|
0.526
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr19_+_42901297
|
0.525
|
|
|
|
|
chr6_-_26199439
|
0.523
|
NM_021065
NM_003530
|
HIST1H2AD
HIST1H3D
|
histone cluster 1, H2ad
histone cluster 1, H3d
|
|
chr22_+_19701968
|
0.523
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr7_-_77325539
|
0.521
|
|
|
|
|
chr19_-_14228362
|
0.521
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr4_-_84035888
|
0.519
|
|
PLAC8
|
placenta-specific 8
|
|
chr16_+_4743709
|
0.518
|
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr1_-_150602047
|
0.518
|
|
ENSA
|
endosulfine alpha
|
|
chr6_+_31126114
|
0.517
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|